# installs
! pip install -q duckdb
! pip install -q mongita
! pip install -q kuzu
! pip install -q lancedb
! pip install -q sentence-transformers
! pip install -q requestsIn my teaching, I often want to demonstrate a variety of database options to my students. However, setting up servers introduces a layer of complexity that isn’t practical for certain offerings. Fortunately, there’s a category of databases that eliminate the need for running servers. These are commonly referred to as embedded, or in-process databases.
Below, I’ll highlight some options across different database types:
- SQLite: A lightweight OLTP database.
- DuckDB: A fast OLAP database quickly becoming a favorite tool for data professionals.
- Mongita: A document database designed to mimic MongoDB’s functionality.
- Kuzu: A graph database that leverages Neo4j’s Cypher query language.
- LanceDB: A vector database built for modern data applications.
Let’s get started.
Setup
# imports
import pandas as pd
import requests
from sentence_transformers import SentenceTransformer
# get an example database for sqlite
! wget -qO music.sqlite https://github.com/lerocha/chinook-database/raw/refs/heads/master/ChinookDatabase/DataSources/Chinook_Sqlite.sqlite
# get an example for the graph database
! wget -q https://files.grouplens.org/datasets/movielens/ml-latest-small.zipI am on a Mac and installed wget via homebrew. To follow along, the easiest path might be to follow along via Google Colab.
With the libraries installed and some sample data downloaded from the web, let’s define the connections to each database.
# instantiate each database
# oltp - single file
import sqlite3
oltp = sqlite3.connect("music.sqlite")
# olap - single file
import duckdb
olap = duckdb.connect("olap.duckdb")
# doc - creates a folder
from mongita import MongitaClientDisk
doc = MongitaClientDisk("docdb")
# graph - creates a folder
import kuzu
g = kuzu.Database("graphdb")
graph = kuzu.Connection(g)
# vector - creates a folder
import lancedb
vec = lancedb.connect("vectordb")OLTP via SQLite
I won’t spend much time on SQlite and python, as there are plenty of tutorials on the web. Below will demonstrate how to interface with sqlite via Python and pandas.
# execute a query to show the tables
# list tables in sqlite database
oltp.execute("SELECT name FROM sqlite_master WHERE type='table';").fetchall()[('Album',),
('Artist',),
('Customer',),
('Employee',),
('Genre',),
('Invoice',),
('InvoiceLine',),
('MediaType',),
('Playlist',),
('PlaylistTrack',),
('Track',)]# of course, we can query data directly into pandas
# join the artists and albumns
sql = """
select a.*,
b.albumid,
b.title
from artist a
join album b
on a.artistid = b.artistid
"""
df = pd.read_sql(sql, oltp)
df.head(3)
| ArtistId | Name | AlbumId | Title | |
|---|---|---|---|---|
| 0 | 1 | AC/DC | 1 | For Those About To Rock We Salute You |
| 1 | 2 | Accept | 2 | Balls to the Wall |
| 2 | 2 | Accept | 3 | Restless and Wild |
# create a silly table
create_table_query = """
CREATE TABLE IF NOT EXISTS applicants (
id INTEGER PRIMARY KEY,
name TEXT,
position TEXT,
salary REAL
)
"""
# Execute the query
oltp.execute(create_table_query)
oltp.commit()Ensure the table exists.
oltp.execute("SELECT name FROM sqlite_master WHERE type='table';").fetchall()[('Album',),
('Artist',),
('Customer',),
('Employee',),
('Genre',),
('Invoice',),
('InvoiceLine',),
('MediaType',),
('Playlist',),
('PlaylistTrack',),
('Track',),
('applicants',)]# a simple dataset
applicants = [
("Alice", "Manager", 70000),
("Bob", "Analyst", 50000),
("Charlie", "Clerk", 30000)
]
# insert into the table,
insert_query = """
INSERT INTO applicants (name, position, salary) VALUES (?, ?, ?)
"""
oltp.executemany(insert_query, applicants)
oltp.commit()# lets grab the data with pandas
pd.read_sql("SELECT * FROM applicants", oltp)| id | name | position | salary | |
|---|---|---|---|---|
| 0 | 1 | Alice | Manager | 70000.0 |
| 1 | 2 | Bob | Analyst | 50000.0 |
| 2 | 3 | Charlie | Clerk | 30000.0 |
# of course we can update records
update_query = """
UPDATE applicants SET salary = ? WHERE name = ?
"""
oltp.execute(update_query, (80000, "Alice"))
oltp.commit()
pd.read_sql("SELECT * FROM applicants", oltp)| id | name | position | salary | |
|---|---|---|---|---|
| 0 | 1 | Alice | Manager | 80000.0 |
| 1 | 2 | Bob | Analyst | 50000.0 |
| 2 | 3 | Charlie | Clerk | 30000.0 |
# we can even use pandas to insert/append to the table
new_data = pd.DataFrame({
"name": ["David", "Eve"],
"position": ["Engineer", "HR"],
"salary": [60000, 55000]
})
new_data.to_sql("applicants", oltp, if_exists="append", index=False)2# review the data
applicants_table = pd.read_sql("SELECT * FROM applicants", oltp)
applicants_table| id | name | position | salary | |
|---|---|---|---|---|
| 0 | 1 | Alice | Manager | 80000.0 |
| 1 | 2 | Bob | Analyst | 50000.0 |
| 2 | 3 | Charlie | Clerk | 30000.0 |
| 3 | 4 | David | Engineer | 60000.0 |
| 4 | 5 | Eve | HR | 55000.0 |
OLAP via DuckDB
I am a huge fan of DuckDB.
- Like SQLite, it’s an in-process database. You can also use duckdb in-memory as well.
- Incredibly performant
- Can be used as an analytics engine or a backend to power data applications.
- Motherduck is a cloud hosted data warehouse based on Duckdb.
There are a variety of applications. If you are interested in learning more about how DuckDB is empowering data teams, I refer you to the free resource here.
# we can create a table directly from pandas
olap.execute("CREATE TABLE applicants as select * from applicants_table")<duckdb.duckdb.DuckDBPyConnection at 0x1453148f0># verify that the data exist
olap.execute("SELECT * FROM applicants").df()| id | name | position | salary | |
|---|---|---|---|---|
| 0 | 1 | Alice | Manager | 80000.0 |
| 1 | 2 | Bob | Analyst | 50000.0 |
| 2 | 3 | Charlie | Clerk | 30000.0 |
| 3 | 4 | David | Engineer | 60000.0 |
| 4 | 5 | Eve | HR | 55000.0 |
# create two csv files
applicants_table.to_csv("e1.csv", index=False)
applicants_table.to_csv("e2.csv", index=False)# duckdb is amazing - scan a directory of files and return as a single dataframe
combined_csvs = olap.execute("SELECT * FROM '*.csv'").df()
combined_csvs| id | name | position | salary | |
|---|---|---|---|---|
| 0 | 1 | Alice | Manager | 80000.0 |
| 1 | 2 | Bob | Analyst | 50000.0 |
| 2 | 3 | Charlie | Clerk | 30000.0 |
| 3 | 4 | David | Engineer | 60000.0 |
| 4 | 5 | Eve | HR | 55000.0 |
| 5 | 1 | Alice | Manager | 80000.0 |
| 6 | 2 | Bob | Analyst | 50000.0 |
| 7 | 3 | Charlie | Clerk | 30000.0 |
| 8 | 4 | David | Engineer | 60000.0 |
| 9 | 5 | Eve | HR | 55000.0 |
Resources
Document Database via Mongita
From the repo, Mongita is a lightweight embedded document database that implements a commonly-used subset of the MongoDB/PyMongo interface.
To demonstrate some basic data operations, I am going to use the Fruityvice API.
# get the data from the API
resp = requests.get("https://www.fruityvice.com/api/fruit/all")
fruits_resp = resp.json()
fruits_resp[0]{'name': 'Persimmon',
'id': 52,
'family': 'Ebenaceae',
'order': 'Rosales',
'genus': 'Diospyros',
'nutritions': {'calories': 81,
'fat': 0.0,
'sugar': 18.0,
'carbohydrates': 18.0,
'protein': 0.0}}# from the documentation
# create the database within the document store
fruits_db = doc.fruits_db# the collection
fruits = fruits_db.fruits# add documents to the fruits collection
fruits.insert_many(fruits_resp)<mongita.results.InsertManyResult at 0x112bea0f0># how many documents are there - no query filter
fruits.count_documents({})49# get one document
fruits.find_one(){'name': 'Persimmon',
'id': 52,
'family': 'Ebenaceae',
'order': 'Rosales',
'genus': 'Diospyros',
'nutritions': {'calories': 81,
'fat': 0.0,
'sugar': 18.0,
'carbohydrates': 18.0,
'protein': 0.0},
'_id': ObjectId('677187a7635da2c06e2f038a')}# we can query documents where the calories key is greater than 100
list( fruits.find({'nutritions.calories': {'$gt': 100}}) )[{'name': 'Durian',
'id': 60,
'family': 'Malvaceae',
'order': 'Malvales',
'genus': 'Durio',
'nutritions': {'calories': 147,
'fat': 5.3,
'sugar': 6.75,
'carbohydrates': 27.1,
'protein': 1.5},
'_id': ObjectId('677187a7635da2c06e2f038f')},
{'name': 'Avocado',
'id': 84,
'family': 'Lauraceae',
'order': 'Laurales',
'genus': 'Persea',
'nutritions': {'calories': 160,
'fat': 14.66,
'sugar': 0.66,
'carbohydrates': 8.53,
'protein': 2.0},
'_id': ObjectId('677187a7635da2c06e2f03ac')},
{'name': 'Hazelnut',
'id': 96,
'family': 'Betulaceae',
'order': 'Fagales',
'genus': 'Corylus',
'nutritions': {'calories': 628,
'fat': 61.0,
'sugar': 4.3,
'carbohydrates': 17.0,
'protein': 15.0},
'_id': ObjectId('677187a7635da2c06e2f03b3')}]Resources
The Github repo is fantastic. Review the README. As you will see, the library is also pretty performant!
Graph Database via Kuzu
Kuzu is an embedded graph database. A graph databases allow us to store our data as nodes and edges, where nodes are the entities in our application, and the edges, or relationships, are how the nodes are related.
While I am a huge fan of Neo4j, Kuzu provides an attractive solution that also aims to focus on graph analytics tasks.
For this demo, I am going to use a small subset Movielens dataset, which was downloaded via wget above.
# unzip the dataset
! unzip ml-latest-small.zipArchive: ml-latest-small.zip
creating: ml-latest-small/
inflating: ml-latest-small/links.csv
inflating: ml-latest-small/tags.csv
inflating: ml-latest-small/ratings.csv
inflating: ml-latest-small/README.txt
inflating: ml-latest-small/movies.csv # read in two of the datasets
movies = pd.read_csv("ml-latest-small/movies.csv")
ratings = pd.read_csv("ml-latest-small/ratings.csv")movies.sample(3)| movieId | title | genres | |
|---|---|---|---|
| 2289 | 3036 | Quest for Fire (Guerre du feu, La) (1981) | Adventure|Drama |
| 9711 | 187717 | Won't You Be My Neighbor? (2018) | Documentary |
| 1753 | 2351 | Nights of Cabiria (Notti di Cabiria, Le) (1957) | Drama |
ratings.sample(3)| userId | movieId | rating | timestamp | |
|---|---|---|---|---|
| 23954 | 166 | 3052 | 3.5 | 1188773616 |
| 42564 | 288 | 2380 | 1.0 | 975692910 |
| 483 | 4 | 3851 | 5.0 | 986849180 |
Kuzu can import from a variety of sources, including Pandas dataframes. For the dataset above, we need to parse out the users into their own dataframe.
# kuzu can read from pandas dataframes
user = ratings[['userId']].drop_duplicates()
user.to_csv("user.csv", index=False)
user.head(3)| userId | |
|---|---|
| 0 | 1 |
| 232 | 2 |
| 261 | 3 |
Unlike Neo4j, it appears that we have define Node and Edge/Relationship tables prior to import. We will do that below.
# create the schema - kuzu specific
# nodes
graph.execute("CREATE NODE TABLE Movie(movieId INT64, title STRING, genres STRING, PRIMARY KEY (movieId))")
graph.execute("CREATE NODE TABLE User(userId INT64, PRIMARY KEY (userId))")
# relationships/edges
graph.execute("CREATE REL TABLE Rating(FROM User TO Movie, rating FLOAT, timestamp INT64)")<kuzu.query_result.QueryResult at 0x1459c6b70># import the data directly from the DataFrames
graph.execute('COPY Movie FROM movies')
graph.execute('COPY User FROM user')
graph.execute('COPY Rating FROM ratings')<kuzu.query_result.QueryResult at 0x1471a2ed0>Below will be a cypher query that queries the graph database via a pattern that looks at user’s rating movies, and then summarizes the data to sort the movies based on the number of ratings while also displaying the average rating, which is a property on the rating relationship.
# query
cql = """
MATCH (u:User)-[r]->(m:Movie)
WITH m.title AS title,
AVG(r.rating) AS avg_rating,
COUNT(r) AS num_ratings
ORDER BY num_ratings DESC
LIMIT 10
RETURN title, avg_rating, num_ratings
"""
graph.execute(cql).get_as_df()
| title | avg_rating | num_ratings | |
|---|---|---|---|
| 0 | Forrest Gump (1994) | 4.164134 | 329 |
| 1 | Shawshank Redemption, The (1994) | 4.429022 | 317 |
| 2 | Pulp Fiction (1994) | 4.197068 | 307 |
| 3 | Silence of the Lambs, The (1991) | 4.161290 | 279 |
| 4 | Matrix, The (1999) | 4.192446 | 278 |
| 5 | Star Wars: Episode IV - A New Hope (1977) | 4.231076 | 251 |
| 6 | Jurassic Park (1993) | 3.750000 | 238 |
| 7 | Braveheart (1995) | 4.031646 | 237 |
| 8 | Terminator 2: Judgment Day (1991) | 3.970982 | 224 |
| 9 | Schindler's List (1993) | 4.225000 | 220 |
Resources
- https://docs.kuzudb.com/import/copy-from-dataframe/
- Learn Cypher, which has been widely adopated as the query language for graph databases.
Vector Database via lancedb
LanceDB is an embeddable vector database, a format that has become incredibly popular with the rise of LLMs and Generative AI.
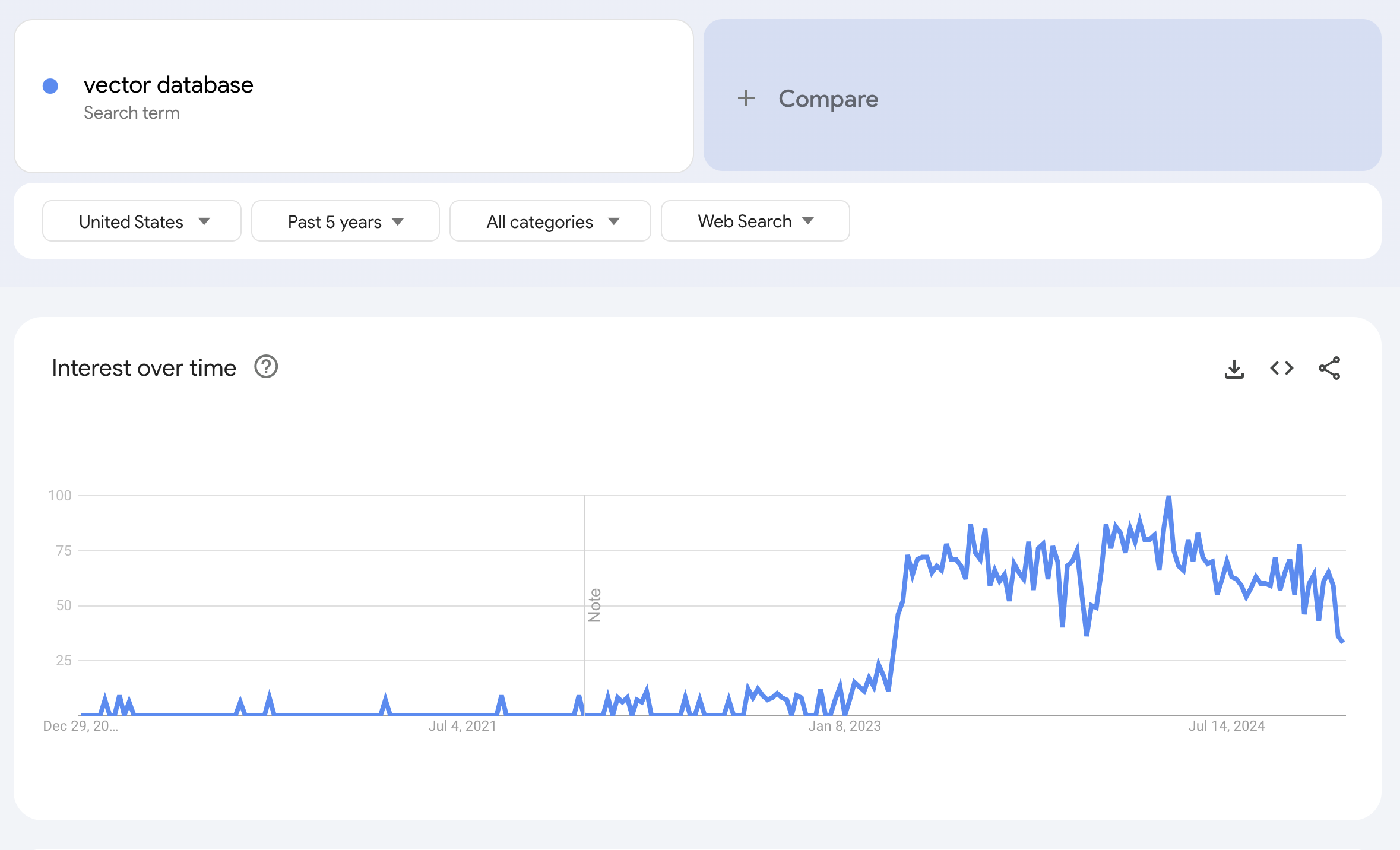
Below is a search of Vector Database over the last 5 years, which spikes just after the release of ChatGPT in November 2022.
# a synthetic dataset of text abstracts
abstracts = [
{
"id": "1",
"title": "Optimizing Transformers for Real-Time Applications",
"abstract": "This paper explores optimization techniques for transformer architectures to reduce latency in real-time applications. We evaluate pruning, quantization, and hardware-specific tuning, achieving up to 40% reduction in inference time without compromising accuracy.",
"topic": "Transformer Optimization"
},
{
"id": "2",
"title": "Adversarial Robustness in Deep Neural Networks",
"abstract": "We present a novel adversarial training algorithm that enhances the robustness of deep neural networks against various attack strategies. Our method outperforms baseline approaches by 15% on benchmark datasets like CIFAR-10 and ImageNet.",
"topic": "Adversarial Robustness"
},
{
"id": "3",
"title": "Generative Adversarial Networks for Data Augmentation",
"abstract": "This study proposes a GAN-based data augmentation technique to improve model generalization. Our experiments demonstrate significant accuracy improvements in medical imaging and natural language processing tasks.",
"topic": "GANs and Data Augmentation"
},
{
"id": "4",
"title": "Explainability in Large Language Models",
"abstract": "We introduce a framework for probing the decision-making processes of large language models, offering insights into token importance and context understanding. Case studies reveal increased transparency in summarization and classification tasks.",
"topic": "Explainability in AI"
},
{
"id": "5",
"title": "Efficient Training of Sparse Neural Networks",
"abstract": "Our research focuses on sparse neural networks and their potential for reducing computational overhead. Using structured pruning techniques, we achieve up to 60% parameter reduction while maintaining competitive performance.",
"topic": "Sparse Neural Networks"
}
]# initialize an embedding model
model = SentenceTransformer("all-MiniLM-L6-v2")# apply the model the abstracts
for a in abstracts:
a["vector"] = model.encode(a["abstract"])# create a table index
tbl = vec.create_table("abstracts", data=abstracts)# database inspection
vec.table_names()['abstracts']# get the embedding for a new abstract
new_abstract = [
{
"id": "20",
"title": "Automating Feature Engineering with Generative AI",
"abstract": "We propose a generative AI framework for automating feature engineering in machine learning pipelines. Case studies demonstrate reduced development time and improved model performance.",
"topic": "Automated Feature Engineering"
}
]
## nn search
embedding = model.encode(new_abstract[0]["abstract"])
# search
results = tbl.search(embedding).limit(3).to_pandas()
results| id | title | abstract | topic | vector | _distance | |
|---|---|---|---|---|---|---|
| 0 | 3 | Generative Adversarial Networks for Data Augme... | This study proposes a GAN-based data augmentat... | GANs and Data Augmentation | [-0.05354594, -0.09120101, 0.08235424, 0.03139... | 1.060543 |
| 1 | 2 | Adversarial Robustness in Deep Neural Networks | We present a novel adversarial training algori... | Adversarial Robustness | [-0.09800007, -0.018522585, -0.0128599, 0.0551... | 1.359992 |
| 2 | 1 | Optimizing Transformers for Real-Time Applicat... | This paper explores optimization techniques fo... | Transformer Optimization | [-0.06164691, 0.058088887, -0.047632158, -0.01... | 1.428792 |
Resources
- The documentation is fantastic and provide a range of resources, including conceptual understanding of where Vector databases fit into the analytics ecosystem.
Summary
Above is a brief review of how to get started with a variety of databases that can power application and analytical workloads, especially those that are targeted at embedded applications that do not require a client/server architecture.
It’s worth noting that there are online hosted versions of the technologies above. The list below highlights offerings that have generous free tiers, which have proven to be helpful for in-class projects.